Pathbase
Pathbase was originally funded under the European Commission's
Fifth framework programme
(QLRI-CT-1999-00320) as a thematic network with the aim of providing a
community
resource for transgenic and mutant rodent pathology.
Its two key aims are:
- To provide a searchable database of histopathology images derived from experimental manipulation of the mouse genome or experiments conducted on genetically manipulated mice.
- A reference / didactic resource covering all aspects of mouse pathology
Participants
Co-ordinator
Dept. of Anatomy, University of Cambridge, Downing
Street, Cambridge, CB2
3DY, UK.
The Pathbase Consortium
The Pathbase Consortium is made up of a group of Pathologists and Geneticists from across Europe and the USA located in the following Laboratories:
Overall objectives of the project
Transgenic rodents are a key tool in understanding human and Analysis of these systems requires considerable expertise in pathology and it is the aim of this project to integrate this expertise in Europe and to teach and inform scientists using transgenic rodents.
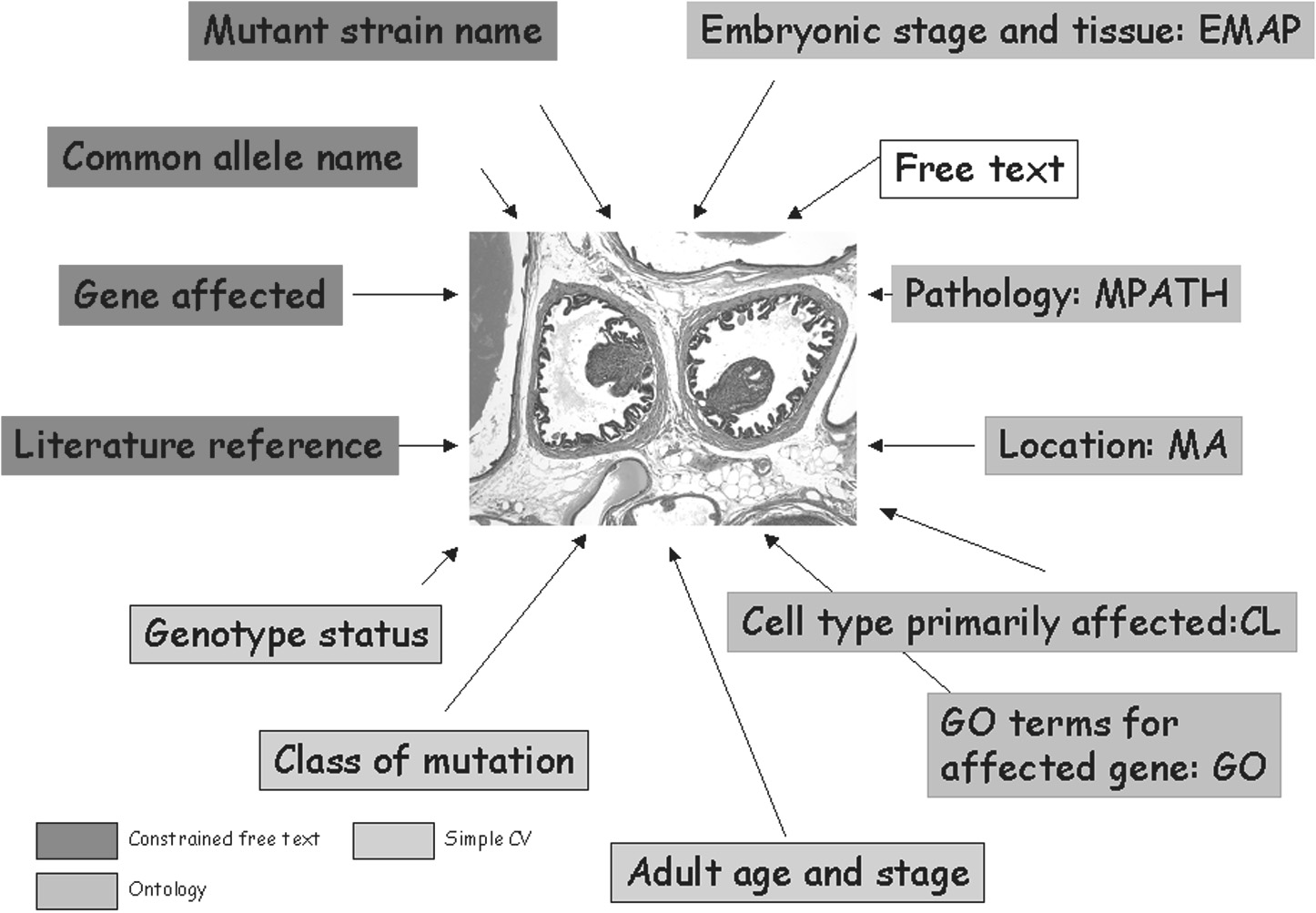
Pathbase is a database that stores images of the abnormal histology associated with spontaneous and induced mutations of both embryonic and adult mice including those produced by transgenesis, targeted mutagenesis and chemical mutagenesis.
Images of normal mouse histology and strain-dependent background lesions are also available. The database and the images are publicly accessible (http://www.pathbase.net) and linked by anatomical site, gene and other identifiers to relevant databases; there are also facilities for public comment and record annotation.
The database is structured around a novel ontology of mouse disorders (MPATH) and provides high-resolution downloadable images of normal and diseased tissues that are searchable through orthogonal ontologies for pathology, developmental stage, anatomy and gene attributes (GO terms), together with controlled vocabularies for type of genetic manipulation or mutation, genotype and free text annotation for mouse strain and additional attributes.
The
database is actively curated and data records assessed by pathologists
in the Pathbase Consortium before publication. The database interface
is designed to have optimal browser and platform compatibility and to
interact directly with other web-based mouse genetic resources.
Experimental approach and working method
Pathbase has put together a consortium of pathologists and veterinarians from across Europe and the USA to set up and run the database. The members have a wide variety of expertise, including radiopathology, to cover the key areas in which we are inviting submissions. Further specialists are called upon when necessary.
The database is accessible on http://www.pathbase.net. Researchers are able to upload their own images onto the database, using a browser based console, with keywords, descriptions, comments, and cross-references to other databases and the literature. Advice may be given on these submissions from a team of Pathologists, members of the project, which may be of assistance to the originator. In addition, feedback forms are provided so that other users can add comments on particular images and diagnoses. This provides a great deal of potentially invaluable interactivity with the database, which many researchers will welcome.
Achievements and results to date
We have developed software for the running and maintenance of the database as a resource for the scientific community. We have now collected together a database of images which will be used both in the database but also on a site designed to teach and inform those using mutant and inbred laboratory mice for biomedical research.
Submissions are made by individual users through web interfaces and although these are checked by the Pathology Committee, who may question diagnoses or provide more information; the overall accuracy of the input is the responsibility of the submitting laboratory.
The Pathology Consortium has developed controlled vocabularies
and an ontology
(MPATH) which cover the whole of general pathology, tissue anatomy and
commonly used
mouse strains. The aim of these vocabularies is to act as functional
descriptors
which optimise searching efficiency rather than as a new detailed
systematic
pathology. The Pathology ontology can be found here
and is posted on the
Gene Ontology Consortium's web site here.
Data may be submitted directly as JPGs or TIFFs with automatic processing and generation of thumbnail images and data report pages. We can accept images up to 8Mb at 300 pixels per inch resolution or higher. The Committee will also undertake a limited amount of digital photography for users who wish to send slides, and can also scan transparencies and prints. We aim to provide assistance with diagnoses as much as is feasible.
The data forms returned from a search contain a feedback field
for other
users to upload comments or to contact the orginator. In this way the
database
becomes a true community resource and allows different laboratories to
compare
data and make contact with each other. Initial trialling of this
facility
suggests that this will be a very valuable way of pooling expertise.
More details on the data structure, implementation of ontologies and development of the database can be found in the following publications:
Schofield, P. N., J. B. Bard, et al. (2004). "Pathbase: a new reference resource and database for laboratory mouse pathology." Radiat Prot Dosimetry 112(4): 525-8.

The Pathbase Consortium is working with EUMORPHIA in the development of a mutant mouse phenotyping platform for mouse pathology details of which can be found on the EMPReSS web site.

Current Status
The database currently holds more than 1000 images of lesions
from mutant mice and their inbred backgrounds and further images are
being added continuously.
A Guide to the mouse necropsy with useful links is now
available on http://eulep.pdn.cam.ac.uk/Necropsy_of_the_Mouse/index.php
and we have recently added a guide to the pathology of mouse skin and
hair mutants, Skinbase,
funded by the North
American Hair Research Society.
How to access Pathbase?
Please go to 'How to use Pathbase' on the menu at the bottom of this page.
Future developments of Pathbase will be posted here and on the Jackson Laboratory's mgi list http://lists.informatics.jax.org/lists/

